
Ava Udvadia
Education
- Postdoctoral Fellow, Neuroscience, Duke University, 1996-2001
- PhD, Molecular Cancer Biology, Duke University, 1995
- BS, Cell and Molecular Biology, University of Michigan, 1989
Research Interests
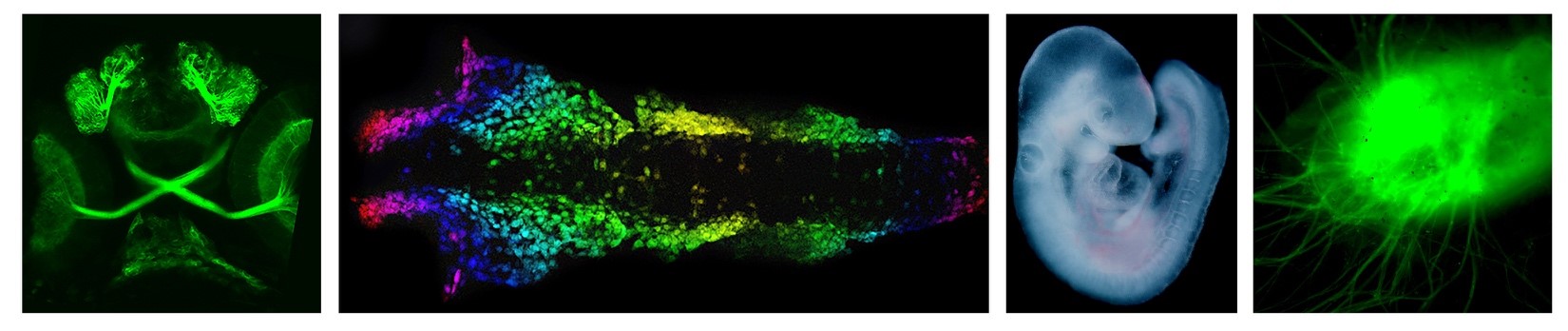
The broad focus of the Udvadia lab is on cellular programming that governs nervous system development and regeneration. More specifically, we use fish and rodent models to understand how dynamic changes in transcription factor activity, chromatin accessibility and chromatin architecture govern:
- Reprogramming of adult retinal ganglion cell neurons for successful CNS
 optic nerve regeneration. In humans, optic nerve damage caused by trauma or optic neuropathies such as glaucoma can result in permanent visual loss. The permanence of the damage is due to a failure of the human central nervous system (CNS) to support regenerative nerve growth. In contrast, fish naturally respond to optic nerve injury by re-establishing the growth capacity of CNS neurons and eventually recovering visual function. It is well known that the cellular programming regulating nerve growth and guidance in the developing visual system is highly conserved between fish and humans. Once the mature visual circuitry has been established, the cells undergo a change in cellular programming from a growth state to one that enables neurotransmission. Thus, what appears to set fish apart from humans is their ability to reprogram adult retinal neurons for wiring in response to optic nerve injury. We hypothesize that discrete evolutionary changes in regeneration-associated gene regulatory interactions in mammals disrupt an otherwise evolutionarily conserved program for functional regeneration of RGC axons. Our current research in this area focuses on: (1) Changes in 3D chromatin architecture that enable regenerative reprogramming, and (2) Dynamic changes in transcription factor interactions that drive stage-specific regeneration-associated gene expression. The long-term goal of this part of our research program is to elucidate key regulatory interactions missing in the mammalian response to optic nerve injury when compared to that of zebrafish in order to discover therapeutic approaches for promoting functional optic nerve regeneration in humans that can restore visual function.
optic nerve regeneration. In humans, optic nerve damage caused by trauma or optic neuropathies such as glaucoma can result in permanent visual loss. The permanence of the damage is due to a failure of the human central nervous system (CNS) to support regenerative nerve growth. In contrast, fish naturally respond to optic nerve injury by re-establishing the growth capacity of CNS neurons and eventually recovering visual function. It is well known that the cellular programming regulating nerve growth and guidance in the developing visual system is highly conserved between fish and humans. Once the mature visual circuitry has been established, the cells undergo a change in cellular programming from a growth state to one that enables neurotransmission. Thus, what appears to set fish apart from humans is their ability to reprogram adult retinal neurons for wiring in response to optic nerve injury. We hypothesize that discrete evolutionary changes in regeneration-associated gene regulatory interactions in mammals disrupt an otherwise evolutionarily conserved program for functional regeneration of RGC axons. Our current research in this area focuses on: (1) Changes in 3D chromatin architecture that enable regenerative reprogramming, and (2) Dynamic changes in transcription factor interactions that drive stage-specific regeneration-associated gene expression. The long-term goal of this part of our research program is to elucidate key regulatory interactions missing in the mammalian response to optic nerve injury when compared to that of zebrafish in order to discover therapeutic approaches for promoting functional optic nerve regeneration in humans that can restore visual function.
- Programming decisions for ectodermal and mesoectodermal cell fate choices during cranial neural crest cell differentiation.
 Disruption of cranial neural crest (CNC) cell development can manifest as a wide variety of human congenital birth defects, including malformations of the craniofacial skeleton, and can also lead to the formation of aggressive tumors, such as neuroblastoma. Contributing to the diversity of anomalies is the ability of the CNC to give rise to both ectodermal cell types, such as sensory ganglia and peripheral glia, and mesectodermal cell types, including the cartilage, bone, and connective tissues of the face. Underlying this remarkable potential is a temporal cascade of events that regulate CNC development from specification to migration and, finally, lineage diversification - a multistep process where CNC progenitors progressively acquire characteristics necessary for cell type-specific function. Our current research in this area is focused on: (1) elucidating the temporally-regulated gene regulatory modules governing CNC differentiation, and (2) investigating the role of Cabin1, a regulator of transcription factors in the MEF2 and NFAT family, in regulating CNC differentiation. The long-term goal of this part of our research program is to discover molecular targets of genetic and environmental factors that lead to CNC-related malformations, and developing potential therapeutic treatments.
Disruption of cranial neural crest (CNC) cell development can manifest as a wide variety of human congenital birth defects, including malformations of the craniofacial skeleton, and can also lead to the formation of aggressive tumors, such as neuroblastoma. Contributing to the diversity of anomalies is the ability of the CNC to give rise to both ectodermal cell types, such as sensory ganglia and peripheral glia, and mesectodermal cell types, including the cartilage, bone, and connective tissues of the face. Underlying this remarkable potential is a temporal cascade of events that regulate CNC development from specification to migration and, finally, lineage diversification - a multistep process where CNC progenitors progressively acquire characteristics necessary for cell type-specific function. Our current research in this area is focused on: (1) elucidating the temporally-regulated gene regulatory modules governing CNC differentiation, and (2) investigating the role of Cabin1, a regulator of transcription factors in the MEF2 and NFAT family, in regulating CNC differentiation. The long-term goal of this part of our research program is to discover molecular targets of genetic and environmental factors that lead to CNC-related malformations, and developing potential therapeutic treatments.
Selected Publications
Andrea Rau, Sumona P. Dhara, Ava J. Udvadia, Paul L. Auer, 2019. Regeneration Rosetta: an interactive web application to explore regeneration-associated gene expression and chromatin accessibility. G3, doi: 10.1534/g3.119.400729.
Sumona P. Dhara, Andrea Rau, Michael J. Lister, Nicole M. Recka, Michael D. Laiosa, Paul L. Auer, Ava J. Udvadia, 2019. Cellular reprogramming for successful CNS axon regeneration is driven by a temporally changing cast of transcription factors. Scientific Reports, doi: 10.1038/s41598-019-50485-6.
Seema Das, Virinchipuram S. Sreevidya, Ava J. Udvadia, Gyaneshwar Prasad, 2019. Dopamine-induced sulfatase and its regulator are required for Salmonella enterica serovar Typhimurium pathogenesis. Microbiology 165: 302-310.
Maria R. Replogle, Virinchipuram S. Sreevidya, Vivian M. Lee, Michael D. Laiosa, Kurt R. Svoboda, Ava J. Udvadia, 2018. Establishment of a Murine culture system for modeling the temporal progression of cranial and trunk neural crest cell differentiation. Disease Models & Mechanisms, doi: 10.1242/dmm.035097.
Ryan R. Williams, Ishwariya Venkatesh, Damien D. Pearse, Ava J. Udvadia, and Mary Bartlett Bunge, 2015. MASH1/Ascl1a leads to GAP43 expression and axon regeneration in the adult CNS. PLOS One 103: e0118918. (Conceptualization: 50%; Experiments: 5%; Analysis: 50%; Writing: 25%; Ishwariya Venkatesh earned her PhD in my lab)
Evdokia Menelaou, Ava J. Udvadia, Robert L. Tanguay, and Kurt R. Svoboda, 2014. Activation of α2A-containing nicotinic acetylcholine receptors mediates nicotine-induced motor output in embryonic zebrafish. European Journal of Neuroscience 40: 2225-2240.
Kerry N. Veth, Jason R. Willer, Ross F. Collery, Matthew P. Gray, Gregory B. Willer, Daniel S. Wagner, Mary C. Mullins, Ava J. Udvadia, Richard S. Smith, Simon W.M. John, Ronald G. Gregg, Brian A. Link, 2011. Mutations in zebrafish lrp2 result in adult onset ocular pathogenesis that models myopia and other risk factors for glaucoma. PLOS Genetics 7: e1001310.
Dena R. Hammond and Ava J. Udvadia, 2010. Cabin1 expression suggests roles in neuronal development. Developmental Dynamics 239: 2443–2451.
Brandon W. Kusik, Dena R. Hammond, and Ava J. Udvadia, 2010. Transcriptional regulatory regions of gap43 needed in developing and regenerating retinal ganglion cells. Developmental Dynamics 239: 482-495.
Angela Schmoldt, Jennifer Forecki, Dena R. Hammond, and Ava J. Udvadia, 2009. Exploring differential gene expression in zebrafish to teach basic molecular biology skills. Zebrafish 6:187-199.
Ava J. Udvadia, 2008. 3.6 kb genomic sequence from Takifugu capable of promoting axon growth associated gene expression in developing and regenerating zebrafish neurons. Gene Expression Patterns 8: 382-388.
Brandon W. Kusik, Michael J. Carvan III, Ava J. Udvadia, 2008. Detection of mercury in aquatic environments using EPRE reporter zebrafish. Marine Biotechnology, 10: 750-757.
Daniel N. Weber, Victoria Connaughton, John Dellinger, David Klemer, Ava Udvadia, Michael J. Carvan III, 2008. Selenomethionine reduces visual deficits due to developmental methylmercury exposures. Physiology and Behavior 93 (1), p.250-260.
J. Rudi Strickler, Ava J. Udvadia, John Marino, Nick Radabaugh, Josh Ziarek, and Ai Nihongi, 2005. Visibility as a Factor in the Copepod - Planktivorous Fish Relationship. Scientia Marina, 69 (Supp. 1) 111-124.
Elwood Linney and Ava J. Udvadia, 2004. Construction and detection of fluorescent germ-line transgenic zebrafish. Methods in Molecular Biology 254: 271-288.
Ava J. Udvadia and Elwood Linney, 2003. Windows into Development: Historic, Current and Future Perspectives on Transgenic Zebrafish. Developmental Biology, 256: 1-17.
Ava J. Udvadia, Reinhard W. Köster, and J. H. Pate Skene, 2001. GAP-43 Promoter Elements in Transgenic Zebrafish Reveal a Difference in Signals for Axon Growth During CNS Development and Regeneration. Development, 128: 1175-1182.