The purpose of the laboratory is to provide equipment and support to monitor the thermodynamic signatures of biochemical reactions, focusing mainly on the interaction of drugs, or drug candidates, and their biological targets. All such molecular interactions can be characterized by their:
- Binding constant (Ka)
- Enthalpy change (ΔH)
- Entropy change (ΔS)
- Stoichiometry (n)
The dissociation constant, Kd, is defined as 1/Ka whereas Gibbs Free Energy (ΔG) relates directly to the binding constant and is related to changes in entropy and enthalpy:
ΔG = -RTLnKa
ΔG = ΔH – TΔS
The center is housed in Chemistry Room 424 and contains:
- TA Instruments Nano ITC Low Volume System
- TA Instruments Nano DSC System
- TA instruments Complete Degassing Station.
TA Instruments Nano ITC

Isothermal titration calorimetry (ITC) is one of the most sensitive and accurate methods for monitoring ligand binding that does not require labeling or immobilization. ITC is widely used in drug discovery, target validation, and structure activity relationship studies. ITC measures the absorption or generation of heat when a known volume of ligand is injected into a protein solution. The integrated areas under each injection peak are plotted against the molar ratio of the active species to directly determine enthalpy change (ΔH), binding constant (Ka) and stoichiometry (n).
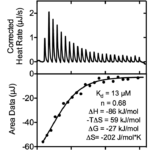
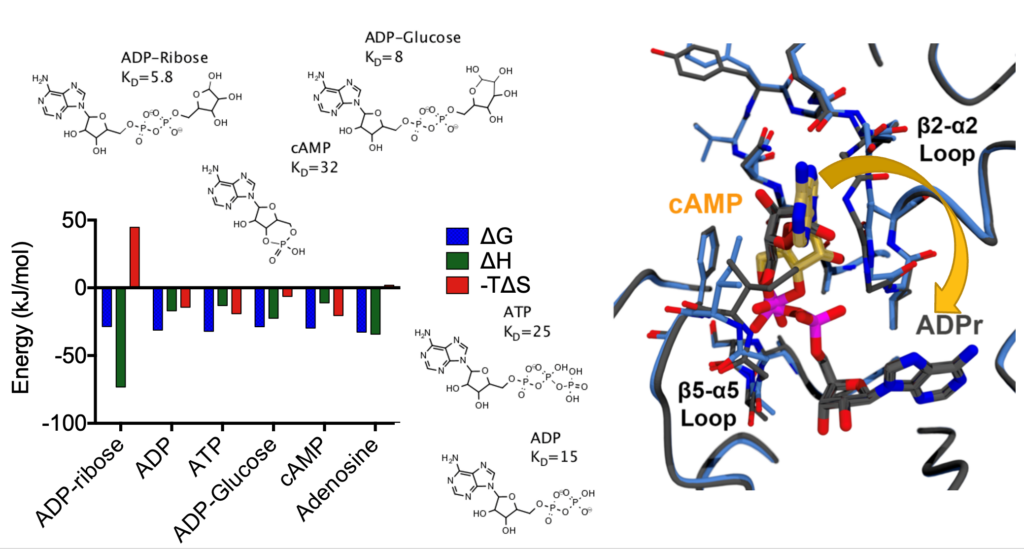
(above) ITC analysis of the SARS-CoV-2 nsp3 Mac1 domain binding to various nucleotides. (Left) Purified Mac1 was titrated with a suspected natural ligand, ADP ribose. (Center) Similar ITC experiments were repeated with each of the compounds listed. (Right) X-ray crystal structure of Mac1 bound to either cAMP (yellow) or ADP-ribose (gray). Thermodynamic differences likely reflect differences in protein-ligand interactions. Note that only ADP ribose binds in a reaction driven by enthalpy and in a conformation where the purine base is uniquely buried.
TA Instruments Nano DSC

Differential scanning calorimetry (DSC) is another valuable tool to study the binding between a biological macromolecule and a drug. DSC is designed for ultra-sensitive measurements of heat absorbed or released by dilute solutions of biomolecules as they are heated or cooled. Unlike ITC, DSC correlates thermodynamics that drive binding (i.e., heat capacity & enthalpy of transition) to changes in the macromolecule caused by the binding reaction. DSC also allows characterization of binding reactions that are incompatible with the stricter requirements of ITC experiments.
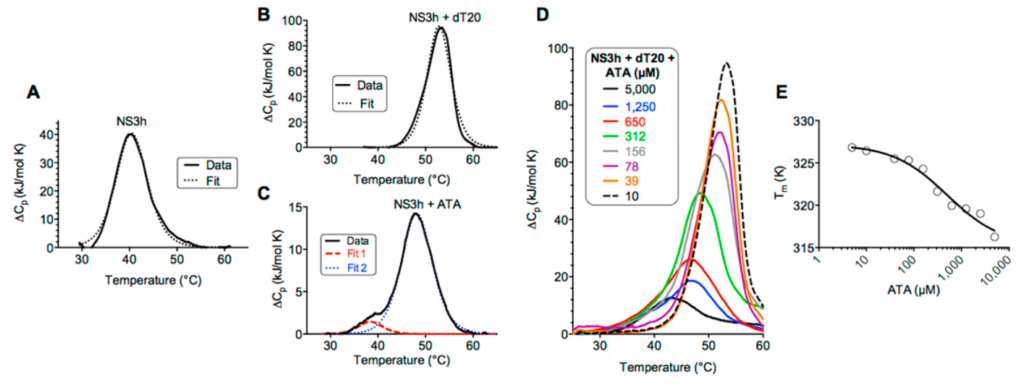
(above) DSC analysis of the hepatitis C virus helicase binding a natural ligand (DNA) or a drug candidate (ATA). (A) Protein was heated from 20 to 100 °C at a rate of 1 °C/min and fitted to a scaled van’t Hoff two-state model (dotted line). (B) Protein and RNA were heated. (C) Protein and ATA were heated. (D) Protein, RNA, and indicated concentrations of ATA were heated. (E) Melting temperatures observed at each ATA concentration for data shown in panel D fitted to a dose response equation.
Degassing Station
The Degassing Station is a stand-alone, touchscreen operated multi-featured accessory for sample preparation and cell cleaning of the Nano ITC and Nano DSC instruments. The degassing chamber accommodates a variety of sample tubes and vials and serves as a stirring platform for effectively degassing samples prior to an ITC or DSC experiment.
Contact: David N. Frick (frickd@uwm.edu)
Sample Publications
Shadrick, W. R., Mukherjee, S., Hanson, A. M., Sweeney, N. L. and Frick, D. N. (2013) Aurintricarboxylic acid modulates the affinity of hepatitis C virus NS3 helicase for both nucleic acid and ATP. Biochemistry 52, 6151-6159. PMC4272346
Frick, D. N., Virdi, R. S., Vuksanovic, N., Dahal, N. and Silvaggi, N. R. (2020) Molecular Basis for ADP-Ribose Binding to the Mac1 Domain of SARS-CoV-2 nsp3. Biochemistry 59, 2608-2615. PMC7341687
Virdi, R. S., Bavisotto, R. V., Hopper, N. C., Vuksanovic, N., Melkonian, T. R., Silvaggi, N. R. and Frick, D. N. (2020) Discovery of Drug-Like Ligands for the Mac1 Domain of SARS-CoV-2 Nsp3. SLAS Discov 25, 1162-1170. PMC7684785